About BRCA1 and Breast Cancer
Breast Cancer
According to breastcancer.org, 1 in 8 women will develop breast cancer in their lifetime. SEER data estimates the incidence of new breast cancer cases (hereditary and nonhereditary) to be 126.1 women in every 100,000 women each year in the US. With such high numbers of individuals developing breast cancer in this country, as well as all over the world, it is important to understand the causes of the disease in order to better diagnose and treat it.
Breastcancer.org also indicates that 20-30% of women diagnosed with breast cancer have members of their family who have also been diagnosed. Only about 5-10% of all cases are germline mutations, that is, the individual inherits a mutation in a gene from their mother or father. Mutations in many genes can predispose someone to breast cancer, but the most common two that are inherited are BRCA1 and BRCA2. BRCA1 is a gene associated with human breast and ovarian cancer susceptibility, which is where its full name comes from: breast cancer-1, early onset. Mutations in this gene are found in familial, early-onset cases of breast cancer. Current estimates on the number of inherited breast cancer cases related to BRCA1 mutations are approximated at 40%, and number of inherited breast and ovarian cancers together are approximated at 80% (Entrez). Though the frequency of BRCA1 mutations varies somewhat between populations and races, it has nonetheless been found in a diverse set of populations. Ashkenazi Jews have the highest incidence of BRCA1 mutations. However, since the overall incidence of mutations in BRCA1 are generally very low, genetic screening for these mutations is not practical, except in cases where there is a strong family history. [1]
Below is a video describing one family's experience with hereditary breast cancer related to BRCA1 mutation.
From YouTube, 2009. The Birds of Hope. Retrieved from http://www.youtube.com/watch?v=KULl_AUwGAU.
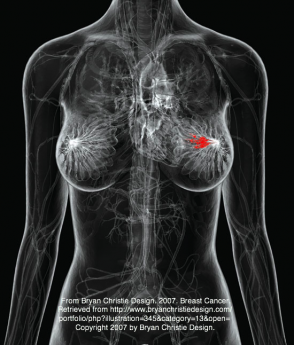
Breast cancer is defined as an uncontrolled growth of mammary tissue, as shown in red in the figure at the top of the page. The disease occurs primarily in females, but is also evident in a small number of males as well. Mammograms are commonly used to detect lumps in the breast before they cause other symptoms. Due to close proximity of the mammary tissue to lymph nodes, these tumors metastasize relatively quickly. Survival rates after 5 years for breast cancer that has not metastasized to the lymph nodes is nearing 98% with treatment, but unfortunately this drops to 27% if the cancer has already established distant metastases. The need for better early detection is evident. Scientists are hoping that by elucidating the mechanisms BRCA1 utilizes to protect cells from growing uncontrolably will aid in the detection and treatment of this cancer. [1]
BRCA1
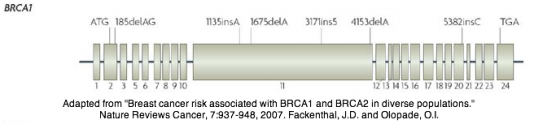
BRCA1 itself is a tumor suppressor gene, and acts as a stabilizing factor for the genome (Entrez). This is evidenced in that approximately 75% of mutations in BRCA1 associated with breast and ovarian cancer are truncations [7]. The wild-type gene is 81,155 base pairs long. The mature mRNA transcript has a varying number of exons due to alternative splicing, but there is a total of 23 exons found in the gene, with exon 11 being the largest. It is important to note, however, that exon 4 is missing, owing to error in the intitial description of the gene [3].
BRCA1 is thought to play a role in localizing DNA damage proteins, including RAD50, MRE11 and p95, to the nucleus in response to DNA damage [15]. These proteins are part of a complex known as the BRCA1-associated genome surveillance complex, or BASC. Other components of this complex are implicated in DNA damage detection and signal transduction (Entrez). BRCA1 itself also associates with RNA polymerase II, the main RNA polymerase in eukaryotes, and contains transcription activation domains [12]. Several alternative splicing transcripts have been characterized, and this seems to affect cellular localization of the protein. In most cases of breast and ovarian cancer, BRCA1 localizes to the cytoplasm instead of the nucleus [2].
This website will focus on genetic and proteomic analysis of BRCA1 by comparing mouse and human sequences and proteins, and by looking at the better characterized interactions within the human cell.
BRCA1 Timeline
1950s - Familial cases of breast cancer are beginning to be described.
1980s - Researchers start using linkage studies to try to identify a gene for breast cancer susceptibility.
1990 - The so-called "breast cancer gene" is localized to chromosome 17q21, though the name BRCA1 is not being used yet [6].
December 1992 - American Association of Cancer Research (AACR) conference discusses the mapping of BRCA1 as a hot topic for breast and ovarian cancer research [5].
October, 1994 - BRCA1 gene is located and sequenced [9]. This is the first report of specific mutations in the candidate region associated with familial cases of breast and ovarian cancer.
February, 1996 - Mouse Brca1 gene is found on chromosome 11 by hybridizing probes based on human BRCA1 cDNA [11].
September, 1996 - Linkage analysis attributes 61% of families with breast cancer in the study to mutations in BRCA1 [10].
February 1999 - BRCA1 nuclear foci formation disappears when benign breast tumors progress to high grade carcinomas [14].
September, 1999 - BRCA1 protein's RING domain is found to be ubiquitinated by ubiquitin-conjugating enzymes [8].
April 2001 - A model is proposed for why breast cancer results from mutations in BRCA1 and BRCA2. The model states that during puberty, the female body has a surge of estrogen to promote mammary tissue growth. During this growth, chromosomal rearrangements facilitated by repetitive elements in both genes causes dysregulated growth, causing tumors [13].
February 2003 - Missense mutations in exon 11 are characterized based on location in functionally important regions of the protein. 38 missense mutations are pointed out as being relevant in breast cancer susceptibility based on homology with other species [4].
[1] American Cancer Society. (2008). Cancer Facts & Figures, 2008 [Brochure]. Atlanta, GA: American Cancer Society.
[2] Chen, Y., Chen, C.F., Riley, D.J., Allred, D.C., Chen, P.L., Von Hoff, D., Osborne, C.K., and Lee, W.H. (1995). Aberrant
subcellular localization of BRCA1 in breast cancer. Science, 270(5237):789-791. doi:10.1126/science.270.5237.789.
[3] Fackenthal, J.D., and Olopade, O.I. (2007). Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations.
Nature Reviews Cancer, 7:937-948. doi:10.1038/nrc2054.
[4] Fleming, M.A., Potter, J.D., Ramirez, C.J., Ostrander, G.K., and Ostrander, E.A. (2003). Understanding missense
mutations in the BRCA1 gene: an evolutionary approach. Proc Natl Acad Sci U.S.A., 100(3):1151-1156.
doi:10.1073/pnas.0237285100.
[5] Foulkes, W. (1992). Genetics of cancer. Lancet, 340(8832), 1402. doi:10.1016/0140-6736(92)92579-5.
[6] Hall, J.M., Lee, M.K., Newman, B., Morrow, J.E., Anderson, L.A., Huey, B., King, M. 1990. Linkage of Early-Onset Familial
Breast Cancer to Chromosome 17q21. Science, 250: 1684-1689. doi:10.1126/science.2270482.
[7] Hogervorst, F.B., Cornelis, R.S., Bout, M., van Vliet, M., Oosterwijk, J.C., Olmer, R., Bakker, B., Klijn, J.G., Vasen, H.F,
Meijers-Heijboer, H., Menko, F.H., Cornelisse, C.J., den Dunnen, J.T., Devilee, P., and van Ommen, G.B. (1995). Rapid
detection of BRCA1 mutations by the protein truncation test. Nature Genetics 10:208-212.doi:10.1038/ng0695-208.
[8] Lorick, K.L., Jensen, J.P., Fang, S., Ong, A.M., Hatakeyama, S., Weissman, A.M. (1999). Ring fingers mediate ubiquitin-
conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U.S.A. 98(20):11364-11369.
doi:10.1073/pnas.96.20.11364.
[9] Miki, Y., Swensen, J., Shattuck-Eidens, D., Futreal, P.A., Harshman, K., Tavtigian, S., Liu, Q., Cochran, C., Bennett, L.M.,
Ding, W., Bell, R., Rosenthal, J., Hussey, C., Tran, T., McClure, M., Frye, C., Hattier, T., Phelps, R., Haugen-Strano, A.,
Katcher, H., Yakumo, K., Gholami, Z., Shaffer, D., Stone, S., Bayer, S., Wray, C., Bogden, R., Dayananth, P., Ward, J.,
Tonin, P., Narod, S., Bristow, P.K., Norris, F.H., Helvering, L., Morrison, P., Rosteck, P., Lai, M., Barrett, J.C., Lewis, C.,
Neuhausen, S., Cannon-Albright, L., Goldgar, D., Wiseman, R., Kamb, A., and Skolnick, M.H. (1994). A stong candidate
for the breast and ovarian cancer susceptibility gene BRCA1. Science, 266(5182), 66-71.
doi:10.1126/science.7545954.
[10] Rebbeck, T.R., Couch, F.J., Kant, J., Calzone, K., DeShano, M., Peng, Y., Chen, K., Garber, J.E., and Weber, B.L. (1996).
Genetic heterogeneity in hereditary breast cancer: role of BRCA1 and BRCA2. American Journal of Human Genetics,
59(3):547-553. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=8751855.
[11] Schrock, E., Badger, P., Larson, D., Erdos, M., Wynshaw-Boris, A., Ried, T., and Brody, L. (1996). The murine homolog
of the human breast and ovarian cancer susceptibility gene Brca1 maps to mouse chromosome 11D. Human Genetics,
97(2):256-259. doi:10.1007/BF02265277.
[12] Scully, R., Anderson, S.F., Chao, D.M., Wei, W., Le, L., Young, R.A., Livingston, D.M., and Parvin, J.D. (1997). BRCA1 is
a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci U.S.A., 94(11):5605-5610.
doi:10.1073/pnas.94.11.5605.
[13] Welsch, P.L., and King, M.C. (2001). BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Human
Molecular Genetics, 10(7):705-713. doi:10.1093/hmg/10.7.705.
[14] Wilson, C.A., Ramos, L., Villaseñor, M.R., Anders, K.H., Press, M.F., Clarke, K., Karlan, B., Chen, J., Scully, R.,
Livingston, D., Zuch, R.H., Kanter, M.H., Cohen, S., Calzone, F.J., and Slamon, D.J. (1999). Localization of human
BRCA1 and its loss in high grade, non-inherited breast carcinomas. Nature Genetics, 21(2):236-240.
doi:10.1038/6029.
[15] Zhong, Q., Chen, C.F., Li, S., Chen, Y., Wang, C.C., Xiao, J., Chen, P.L., Sharp, Z.D., and Lee, W.H. (1999). Association
of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science, 285(5428):747-750.
doi:10.1126/science.285.5428.747.