Further directions
Here I will describe areas of research that I believe may be useful for further examining the role of BRCA1 in breast cancers. Though research on BRCA1 has been ongoing for over two decades now, the exact mechanisms of BRCA1 loss in cancers is not well known. Once we understand the underlying mechanisms, this could lead to more personalized treatment of breast cancer, as well as other cancers in which BRCA1 expression is found to be lost.
First, it is still largely unknown how BRCA1 mutations almost exclusively cause breast and ovarian cancers. The protein is expressed in nearly every cell type in the body, and presumably is functioning in each cell for DNA damage repair. There are multiple redundant DNA repair pathways, and each cell may rely on different mechanisms for fidelity. I propose that further studies examine the expression of different DNA repair pathways in a variety of cell types to determine which repair pathways are essential for each lineage. This could elucidate why BRCA1 causes breast and ovarian cancers specifically, as well as provide valuable insight into specific repair mechanisms on a tissue-specific basis. This would greatly aid treatment of a variety of human cancers, as secondary repair mechanisms may be targets for stimulation when primary repair mechanisms are lost.
Second, further characterizing protein modifications that modulate BRCA1 activity would provide clues as to how the function of BRCA1 is controlled and how sporadic breast cancers lose BRCA1 function when mutations in the gene itself are not common. Phosphorylation sites have been described in a number of studies, but how the majority of these modifications affects protein function is yet to be determined. Likewise, ubiquitination of BRCA1 may be another level at which BRCA1 protein expression is controlled. Because one of BRCA1's roles is to ubiquitinate other proteins through the function of its RING finger domain, determining sites of ubiquitination on BRCA1 has proven to be rather difficult, as BRCA1 normally is ubiquitinated in order to transfer the group to other proteins. Determining "normal" sites of ubiquitination versus abnormal sites will provide another level of insight into protein stability. Both phosphorylation and ubiquitination of BRCA1 may be valuable drug targets for the treatment of breast cancer, potentially allowing BRCA1 function to be rescued.
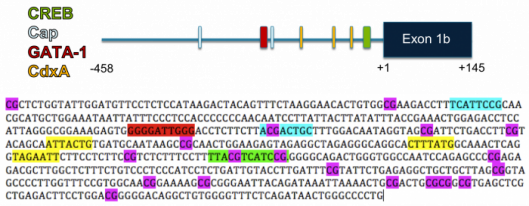
Lastly, DNA methylation has been hypothesized to control the expression of BRCA1, and may be a mechanism by which expression is decreased in sporadic tumors. Promoter methylation is well established to silence genes, and various papers have reported that BRCA1 promoters become methylated in many breast cancer samples. The pattern of methylation of BRCA1 promoters varies from sample to sample, but Mancini, et al. reported that in many cases CpGs within the CREB binding site (CEBP) are methylated, which would prevent CREB from binding the promoter, and possibly limiting transcription from the gene. [1] Below I have made a gene map of the BRCA1 promoter starting 458 positions upstream of the start site. Using MOTIF, I found binding sites within the promoter with high cut-off scores (over 90). These sites are color coded as indicated. Following the gene map, I have color coded the actual sequence to show all CpG sites (in violet), as well as the binding sites (color coded as in map). As you can see, binding sites that contain CpGs, and thus have the potential for disruption through methylation, are only the CREB binding site, as well as the second Cap transcription activation site.
I propose that the role of these transcription activation binding sites should be studied in more detail for BRCA1 and thus should provide more insight into how CpG methylation could affect gene expression. Again there could be a way to target this mechanism with drugs in the future to allow expression, and thus restore DNA repair, in cancers.
[1] Mancini, D.N., Rodenhiser, D.I., Ainsworth, P.J., O'Malley, F.P., Singh, S.M., Xing, W., and Archer, T.K. (1998).
CpG methylation within the 5' regulatory region of the BRCA1 gene is tumor specific and includes a putative
CREB binding site. Oncogene, 16:1161-1169. doi:10.1038/sj.onc.1201630.
Site created by Jessica D. Kueck
Genetics 677 Assignment, Spring 2009
University of Wisconsin-Madison